About bioKepler
With the introduction of the next-generation sequencers, e.g., the 454 Sequencer, there has been a huge increase in the amount of DNA sequence data. These 2nd generation and the emerging 3rd generation sequencing technologies are providing extremely huge data that overwhelms current computational tools and resources.The traditional bioinformatics scenario of downloading data locally to analyze it doesn’t scale at these huge magnitudes. In addition, as datasets get larger, moving data over the network becomes more complicated, error-prone and costly to maintain.
This enormous data growth places unprecedented demands on traditional single-processor bioinformatics algorithms. Efficient and comprehensive analysis of the generated data requires distributed and parallel processing capabilities. Bioinformaticians often conduct parallel computation by dividing their queries or databases to turn them into small jobs to be submitted into computer clusters, Grid or Cloud, and then merge the output. New computational techniques and efficient execution mechanisms for this data-intensive workload are needed. Technologies like scientific workflows and data-intensive computing promise new capabilities to enable rapid analysis of these next-generation sequence data. These technologies, when used together in an integrative architecture, have great promise to serve many projects with similar needs on the emerging distributed data-intensive computing resources.
For enabling bioinformaticians and computational biologists to conduct efficient analysis, there still remains a need for higher-level abstractions on top of scientific workflow systems and distributed computing methods. The bioKepler project is motivated by the following three challenges that remain unsolved:
- How can large-scale sequencing data be analyzed systematically in a way that incorporates and enables reuse of best practices by the scientific community?
- How can such analysis be easily configured or programmed by end users with various skill levels to formulate actual bioinformatics workflows?
- How can such workflows be executed in computing resources available to scientists in an efficient and intuitive manner?
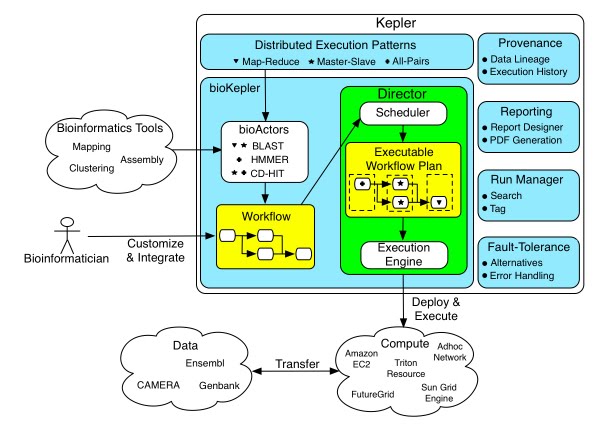
bioKepler is a three year long project that builds scientific workflow components to execute a set of bioinformatics tools using distributed execution patterns. Once customized, these components are executed on multiple distributed platforms including various Cloud and Grid computing platforms. In addition, we deliver virtual machines including a Kepler engine and all bioinformatics tools and applications we are building components for in bioKepler.
The conceptual framework below shows the components of bioKepler module including:
- bioActors that integrate bioinformatics and computational biology tools with distributed patterns;
- inner process of the Distibuted Kepler Director; and
- other modules in Kepler environment, e.g., provenance recorder.
The diagram also shows the computational and data resources the prototype system will be tested on.