Projects
Current Projects
 |
CHASE-CI: Cognitive Hardware and Software Ecosystem Community InfrastructureThis project will build a cloud of hundreds of affordable Graphics Processing Units (GPUs), networked together with a variety of neural network machines to facilitate development of next generation cognitive computing. This cloud will be accessible by 30 researchers assembled from 10 universities via the NSF-funded Pacific Research Platform. These researchers will investigate a range of problems from image and video recognition, computer vision, contextual robotics to cognitive neurosciences using the cloud to be purpose-built in this project. Training of neural network with large data-sets is best performed on GPUs. Lack of availability of affordable GPUs and lack of easy access to the new generation of Non-von Neumann (NvN) machines with embedded neural networks impede research in cognitive computing. The purpose-built cloud will be available over the network to address this bottleneck. PIs will study various Deep Neural Network, Recurrent Neural Network, and Reinforcement Learning Algorithms on this platform. |
IPPD: Integrated end-to-end Performance Prediction and Diagnosis for Extreme Scientific WorkflowsWorkflows are designed to execute on a loosely connected set of distributed and heterogeneous computational resources. It is critically important to have a clear understanding of the factors that influence their performance and for the potential optimization of their execution. The performance of a workflow is determined by a wide range of factors. In IPPD three core issues are being addressed in order to provide insights into workflow execution that can be used to both explain and optimize their execution: 1) provide an expectation of the performance of a workflow in-advance of execution to provide a best baseline performance; 2) identify areas of consistent low performance and diagnose the reason why; and 3) study the important issue of performance variability. The design and analysis of large-scale scientific workflows is difficult precisely because each task can exhibit extreme performance variability. New prediction and diagnostic methods are required to enable efficient use of present and emerging workflow resources. |
 |
 |
WIFIRE: A Scalable Data-Driven Monitoring, Dynamic Prediction and Resilience Cyberinfrastructure for WildfiresThe WIFIRE CI (cyberinfrastructure) builds an integrated system for wildfire analysis, with specific regard to changing urban dynamics and climate. The system integrates networked observations such as heterogeneous satellite data and real-time remote sensor data, with computational techniques in signal processing, visualization, modeling, and data assimilation to provide a scalable method to monitor such phenomena as weather patterns that can help predict a wildfire's rate of spread. Kepler scientific workflows are used in WIFIRE as an integrative distributed programming model and will simplify the implementation of engineering modules for data-driven simulation, prediction and visualization while allowing integration with large-scale computing facilities. |
KeplerThe Kepler Project is dedicated to furthering and supporting the capabilities, use, and awareness of the free and open source, scientific workflow application, Kepler. Kepler is designed to help scientists, analysts, and computer programmers create, execute, and share models and analyses across a broad range of scientific and engineering disciplines. Kepler can operate on data stored in a variety of formats, locally and over the internet, and is an effective environment for integrating disparate software components, such as merging "R" scripts with compiled "C" code, or facilitating remote, distributed execution of models. Using Kepler's graphical user interface, users simply select and then connect pertinent analytical components and data sources to create a "scientific workflow" an executable representation of the steps required to generate results. The Kepler software helps users share and reuse data, workflows, and components developed by the scientific community to address common needs. |
 |
 |
BBDTC: Biomedical Big Data Training Collaborative
The BBDTC is a community-oriented platform to encourage high-quality knowledge dissemination with the aim of growing a well-informed biomedical big data community through collaborative efforts on training and education. The BBDTC is an e-learning platform that empowers the biomedical community to develop, launch and share open training materials through its intuitive user interface. The BBDTC has a growing repository for online materials including lecture videos, PowerPoint presentations, hands-on exercises with real-world examples, and virtual machine toolboxes for biomedical big data activities. The platform also supports course content personalization through the playlist feature to supports biomedical scientists at all levels, and from varied backgrounds. The three primary components of the BBDTC framework are BBDTC User Interface, BBDTC VM Toolbox, and Cross platform integration module. The BBDTC project focuses on continually adding new features to BBDTC framework to enhance user experience as learners and instructors. |
Past Projects
 |
bioKepler: A Comprehensive Bioinformatics Scientific Workflow Module for Distributed Analysis of Large-Scale Biological DatabioKepler project builds a Kepler module to execute bioinformatics tools using distributed execution patterns. Once customized, these components are executed on multiple distributed platforms including various Cloud and Grid computing platforms. In addition, bioKepler delivers virtual machines including a Kepler engine and all bioinformatics tools and applications distributed in bioKepler. bioKepler is a module that is distributed on top of the core Kepler scientific workflow system. |
NBCR: National Biomedical Computation ResourceNBCR enables biomedical scientists to address the challenge of integrating detailed structural measurements from diverse scales of biological organization that range from molecules to organ systems in order to gain quantitative understanding of biological function and phenotypes. Predictive multi-scale models and our driving biological research problems together address issues in modeling of sub-cellular biophysics, building molecular modeling tools to accelerate discovery, and defining tools for patient-specific multi-scale modeling. Kepler is being utilized in NBCR to provide more complete computer and data-aided workflows that are sensible to biological scientists, hide to the extent possible the shifting sands of computing infrastructure as it evolves over years and decades, and are fundamentally sound from the scientific reproducibility viewpoint. |
 |
 |
NIF: The Neuroscience Information FrameworkThe Neuroscience Information Framework is a dynamic inventory of Web-based neuroscience resources: data, materials, and tools accessible via any computer connected to the Internet. An initiative of the NIH Blueprint for Neuroscience Research, NIF advances neuroscience research by enabling discovery and access to public research data and tools worldwide through an open source, networked environment. NIF utilizes Kepler workflows to provide an accumulated view on neuroscience data, e.g., the "Brain data flow", which present a user with categorized information and heat maps about sources which have information on various brain regions. |
Smart Manufacturing Leadership Coalition (SMLC)The Smart Manufacturing Leadership Coalition (SMLC) is a project to build Smart Manufacturing (SM) systems that integrate manufacturing intelligence in real time across an entire production operation including supply chain that are rare in large companies, and virtually non-existent in small and medium size organizations. Even though the sensor, signal and response technologies have become very reliable and advanced, the implementation of data-driven manufacturing intelligence and adoption of real time performance-modeling management in achieving good operating performance across multiple units within the production line of an entire factory has been lagging behind. Implementation of dynamic real time monitoring, analysis and modeling is essential to the development of a sophisticated smart manufacturing intelligence. Workflow and cloud computing are two main components developed in the system to date. SMLC considers workflows as an essential technique in the implementation of automation, and dynamic decision-making process through contextualization and analysis of real time data. Due to the capability to build flexible and complicated applications, each user application service is expressed in a workflow. The cloud platform is intended for elastic scalability and reduces cost of deploying SM services by providing guaranteed compute and data storage resources to complete jobs in real time. |
 |
DISCOSciThe DISCOSci project is an effort to design and build an entirely new type of data collection system for the world's oceans. Within this system, data collection is handled by small, inexpensive, buoyancy-controlled robots called, ‘autonomous underwater explorers’ or AUEs. These AUEs are small enough to ride underwater currents, and record where the ocean takes them. Additional sensors onboard collect important data, such as the current water temperature, or the local concentration of algae. As satellite signals cannot penetrate surface waters, each AUE communicates acoustically with a network of intelligent buoys on the surface. These buoys can relay data and receive instructions in near real-time via satellite from ship or shore. |
 |
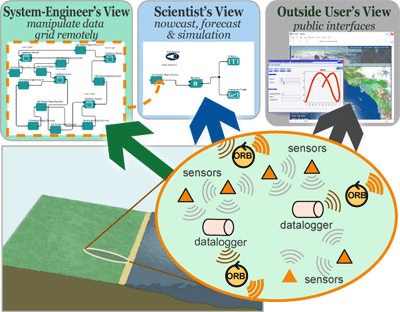 |
Real-time Environment for Analytical Processing (REAP)REAP project investigators are combining the real-time data grid being constructed through other projects (Data Turbine, OPeNDAP, EarthGrid) and the Kepler scientific workflow system to provide a framework for designing and executing scientific workflows that use sensor data. To this end, project collaborators are extending Kepler to access sensor data in workflows, monitor, inspect and control sensor networks, and simulate the design of new sensor networks. |
Ocean Observatories Initiative (OOI)The Ocean Observatories Initiative (OOI) is a program funded by the National Science Foundation to develop a permanent underwater observatory, through the use many different types of sensors, as well as cutting edge network infrastructure. In the past, earlier observations primarily involved ship-based expeditions. OOI's focus is to use new technologies to build a permanent presence in the ocean. OOI's objective is to deliver data and data products, based on physical, chemical, geological, and biological sensor systems, for the next 25 or more years. OOI has been designed with Kepler as an integral component. Kepler workflows are suitable for use in the creation of data products, overseeing quality control, and implementing other systems logic, all with the provenance expected of scientific endeavors. |
 |
 |
Kepler/COREThe Kepler/CORE Project is an NSF-funded effort to coordinate development of Kepler and to enhance the attributes and functions of the system most important for broad adoption and long-term sustainability (see Kepler/CORE Vision & Mission). The primary goal is to serve the broadest set of science communities possible by making the system more comprehensive, open, reliable and extensible. |
Scientific Data Management Center, Scientific Process Automation (SDM Center, SPA)Science in many disciplines increasingly requires data-intensive and compute-intensive information technology (IT) solutions for scientific discovery. Scientific applications with these requirements range from the understanding of biological processes at the sub-cellular level of "molecular machines" (as is common, e.g., in genomics and proteomics), to the level of simulating nuclear fusion reactions and supernova explosions in astrophysics. A practical bottleneck for more effective use of available computational and data resources is often in the IT knowledge of the end-user; in the design of resource access and use of processes; and the corresponding execution environments, i.e., in the scientific workflow environment of end user scientists. The goal of the Kepler/SPA thrust of the SDM Center is to provide solutions and products for effective and efficient modeling, design, configurability, execution, and reuse of scientific workflows. |
 |
 |
CAMERA: Community Cyberinfrastructure for Advanced Microbial Ecology Research and AnalysisCAMERA project serves the needs of the microbial ecology research community, and other scientists using metagenomics data, by creating a rich, distinctive data repository and a bioinformatics tools resource that addresses many of the unique challenges of metagenomic analysis. Kepler scientific workflows are being utilized to launch data analysis tools. Workflows are configurable analysis packages that can be applied to data within the CAMERA workspace or to data uploaded from the local system. |
